 8.3 The Genetic CodeThe genetic code is how a cell translates the information contained in its DNA into amino acids and then proteins, which do the real work in the cell. 8.3.1 BackgroundHerein is a short introduction for the nonbiologists. As stated earlier, DNA encodes the primary structure (i.e., the amino acid sequence) of proteins. DNA has four nucleotides, and proteins have 20 amino acids. The encoding works by taking each group of three nucleotides from the DNA and "translating" them to an amino acid or a stop signal. Each group of three nucleotides is called a codon. We'll see in detail how this coding and translation works. Actually, transcription first uses DNA to make RNA, and then translation uses RNA to make proteins. This is called the central dogma of molecular biology. But in this course, I'll abbreviate the process and somewhat inaccurately call the entire process from DNA to protein "translation." The reason for this cavalier distinction is that the whole business is much easier to simulate on computer using strings to represent the DNA, RNA, and proteins. In fact, as shown in Chapter 4, transcribing DNA to RNA is very easy indeed. In your computer simulations, you can simply skip that step, since it's just a matter of changing one letter to another. (The actual process in the cell, of course, is much more complex.) Note that with four kinds of bases, each group of three bases of DNA can represent as many as 4 x 4 x 4 = 64 possible amino acids. Since there are only 20 amino acids plus a stop signal, the genetic code has evolved some redundancy, so that some amino acids are represented by more than one codon. Every possible three bases of DNA—each codon—represents some amino acid (apart from the three codons that represent a stop signal). The chart in Figure 8-1 shows how the various bases combine to form amino acids. There are many interesting things to note about the genetic code. For our purposes, the most important is redundancy—the way more than one codon translates to the same amino acid. We'll program this using character classes and regular expressions, as you'll soon see.[2]
Figure 8-1. The genetic code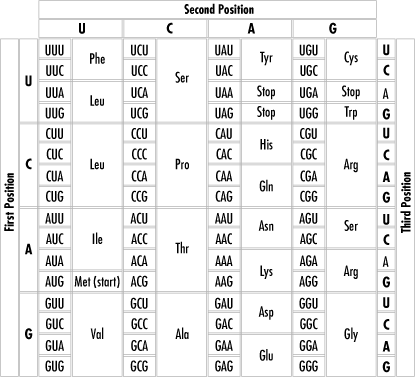 The machinery of the cell actually starts at some point along the RNA and "reads" the sequences codon after codon, attaching the encoded amino acid to the end of the growing protein sequence. Example 8-1 simulates this, reading the string of DNA three bases at a time and concatenating the symbol for the encoded amino acid to the end of the growing protein string. In the cell, the process stops when a codon is encountered. 8.3.2 Translating Codons to Amino AcidsThe first task is to enable the following programs to do the translation from the three-nucleotide codons to the amino acids. This is the most important step in implementing the genetic code, which is the encoding of amino acids by three-nucleotide codons. Here's a subroutine that returns an amino acid (represented by a one-letter abbreviation) given a three-letter DNA codon: # codon2aa
#
# A subroutine to translate a DNA 3-character codon to an amino acid
sub codon2aa {
my($codon) = @_;
if ( $codon =~ /TCA/i ) { return 'S' } # Serine
elsif ( $codon =~ /TCC/i ) { return 'S' } # Serine
elsif ( $codon =~ /TCG/i ) { return 'S' } # Serine
elsif ( $codon =~ /TCT/i ) { return 'S' } # Serine
elsif ( $codon =~ /TTC/i ) { return 'F' } # Phenylalanine
elsif ( $codon =~ /TTT/i ) { return 'F' } # Phenylalanine
elsif ( $codon =~ /TTA/i ) { return 'L' } # Leucine
elsif ( $codon =~ /TTG/i ) { return 'L' } # Leucine
elsif ( $codon =~ /TAC/i ) { return 'Y' } # Tyrosine
elsif ( $codon =~ /TAT/i ) { return 'Y' } # Tyrosine
elsif ( $codon =~ /TAA/i ) { return '_' } # Stop
elsif ( $codon =~ /TAG/i ) { return '_' } # Stop
elsif ( $codon =~ /TGC/i ) { return 'C' } # Cysteine
elsif ( $codon =~ /TGT/i ) { return 'C' } # Cysteine
elsif ( $codon =~ /TGA/i ) { return '_' } # Stop
elsif ( $codon =~ /TGG/i ) { return 'W' } # Tryptophan
elsif ( $codon =~ /CTA/i ) { return 'L' } # Leucine
elsif ( $codon =~ /CTC/i ) { return 'L' } # Leucine
elsif ( $codon =~ /CTG/i ) { return 'L' } # Leucine
elsif ( $codon =~ /CTT/i ) { return 'L' } # Leucine
elsif ( $codon =~ /CCA/i ) { return 'P' } # Proline
elsif ( $codon =~ /CCC/i ) { return 'P' } # Proline
elsif ( $codon =~ /CCG/i ) { return 'P' } # Proline
elsif ( $codon =~ /CCT/i ) { return 'P' } # Proline
elsif ( $codon =~ /CAC/i ) { return 'H' } # Histidine
elsif ( $codon =~ /CAT/i ) { return 'H' } # Histidine
elsif ( $codon =~ /CAA/i ) { return 'Q' } # Glutamine
elsif ( $codon =~ /CAG/i ) { return 'Q' } # Glutamine
elsif ( $codon =~ /CGA/i ) { return 'R' } # Arginine
elsif ( $codon =~ /CGC/i ) { return 'R' } # Arginine
elsif ( $codon =~ /CGG/i ) { return 'R' } # Arginine
elsif ( $codon =~ /CGT/i ) { return 'R' } # Arginine
elsif ( $codon =~ /ATA/i ) { return 'I' } # Isoleucine
elsif ( $codon =~ /ATC/i ) { return 'I' } # Isoleucine
elsif ( $codon =~ /ATT/i ) { return 'I' } # Isoleucine
elsif ( $codon =~ /ATG/i ) { return 'M' } # Methionine
elsif ( $codon =~ /ACA/i ) { return 'T' } # Threonine
elsif ( $codon =~ /ACC/i ) { return 'T' } # Threonine
elsif ( $codon =~ /ACG/i ) { return 'T' } # Threonine
elsif ( $codon =~ /ACT/i ) { return 'T' } # Threonine
elsif ( $codon =~ /AAC/i ) { return 'N' } # Asparagine
elsif ( $codon =~ /AAT/i ) { return 'N' } # Asparagine
elsif ( $codon =~ /AAA/i ) { return 'K' } # Lysine
elsif ( $codon =~ /AAG/i ) { return 'K' } # Lysine
elsif ( $codon =~ /AGC/i ) { return 'S' } # Serine
elsif ( $codon =~ /AGT/i ) { return 'S' } # Serine
elsif ( $codon =~ /AGA/i ) { return 'R' } # Arginine
elsif ( $codon =~ /AGG/i ) { return 'R' } # Arginine
elsif ( $codon =~ /GTA/i ) { return 'V' } # Valine
elsif ( $codon =~ /GTC/i ) { return 'V' } # Valine
elsif ( $codon =~ /GTG/i ) { return 'V' } # Valine
elsif ( $codon =~ /GTT/i ) { return 'V' } # Valine
elsif ( $codon =~ /GCA/i ) { return 'A' } # Alanine
elsif ( $codon =~ /GCC/i ) { return 'A' } # Alanine
elsif ( $codon =~ /GCG/i ) { return 'A' } # Alanine
elsif ( $codon =~ /GCT/i ) { return 'A' } # Alanine
elsif ( $codon =~ /GAC/i ) { return 'D' } # Aspartic Acid
elsif ( $codon =~ /GAT/i ) { return 'D' } # Aspartic Acid
elsif ( $codon =~ /GAA/i ) { return 'E' } # Glutamic Acid
elsif ( $codon =~ /GAG/i ) { return 'E' } # Glutamic Acid
elsif ( $codon =~ /GGA/i ) { return 'G' } # Glycine
elsif ( $codon =~ /GGC/i ) { return 'G' } # Glycine
elsif ( $codon =~ /GGG/i ) { return 'G' } # Glycine
elsif ( $codon =~ /GGT/i ) { return 'G' } # Glycine
else {
print STDERR "Bad codon \"$codon\"!!\n";
exit;
}
}
This code is clear and simple, and the layout makes it obvious what's happening. However, it can take a while to run. For instance, given the codon GGT for glycine, it has to check each test until it finally succeeds on the last one, and that's a lot of string comparisons. Still, the code achieves its purpose. There's something new happening in the code's error message. Recall filehandles from Chapter 4 and how they access data in files. From Chapter 5, remember the special filehandle STDIN that reads user input from the keyboard. STDOUT and STDERR are also special filehandles that are always available to Perl programs. STDOUT directs output to the screen (usually) or another standard place. When a filehandle is missing from a print statement, STDOUT is assumed. The print statement accepts a filehandle as an optional argument, but so far, we've been printing to the default STDOUT. Here, error messages are directed to STDERR, which usually prints to the screen, but on many computer systems they can be directed to a special error file or other location. Alternatively, you sometimes want to direct STDOUT to a file or elsewhere but want STDERR error messages to appear on your screen. I mention these options because you are likely to come across them in Perl code; we don't use them much in this book (see Appendix B for more information). 8.3.3 The Redundancy of the Genetic CodeI've remarked on the redundancy of the genetic code, and the last subroutine clearly displays this redundancy. It might be interesting to express that in your subroutine. Notice that groups of redundant codons almost always have the same first and second bases and vary in the third. You've used character classes in regular expressions to match any of a set of characters. Now, let's try to redo the subroutine to make one test for each redundant group of codons: # codon2aa
#
# A subroutine to translate a DNA 3-character codon to an amino acid
# Version 2
sub codon2aa {
my($codon) = @_;
if ( $codon =~ /GC./i) { return 'A' } # Alanine
elsif ( $codon =~ /TG[TC]/i) { return 'C' } # Cysteine
elsif ( $codon =~ /GA[TC]/i) { return 'D' } # Aspartic Acid
elsif ( $codon =~ /GA[AG]/i) { return 'E' } # Glutamic Acid
elsif ( $codon =~ /TT[TC]/i) { return 'F' } # Phenylalanine
elsif ( $codon =~ /GG./i) { return 'G' } # Glycine
elsif ( $codon =~ /CA[TC]/i) { return 'H' } # Histidine
elsif ( $codon =~ /AT[TCA]/i) { return 'I' } # Isoleucine
elsif ( $codon =~ /AA[AG]/i) { return 'K' } # Lysine
elsif ( $codon =~ /TT[AG]|CT./i) { return 'L' } # Leucine
elsif ( $codon =~ /ATG/i) { return 'M' } # Methionine
elsif ( $codon =~ /AA[TC]/i) { return 'N' } # Asparagine
elsif ( $codon =~ /CC./i) { return 'P' } # Proline
elsif ( $codon =~ /CA[AG]/i) { return 'Q' } # Glutamine
elsif ( $codon =~ /CG.|AG[AG]/i) { return 'R' } # Arginine
elsif ( $codon =~ /TC.|AG[TC]/i) { return 'S' } # Serine
elsif ( $codon =~ /AC./i) { return 'T' } # Threonine
elsif ( $codon =~ /GT./i) { return 'V' } # Valine
elsif ( $codon =~ /TGG/i) { return 'W' } # Tryptophan
elsif ( $codon =~ /TA[TC]/i) { return 'Y' } # Tyrosine
elsif ( $codon =~ /TA[AG]|TGA/i) { return '_' } # Stop
else {
print STDERR "Bad codon \"$codon\"!!\n";
exit;
}
}
Using character classes and regular expressions, this code clearly shows the redundancy of the genetic code. Also notice that the one-character codes for the amino acids are now in alphabetical order. A character class such as [TC] matches a single character, either T or C. The . is the regular expression that matches any character except a newline. The /GT./i expression for valine matches GTA, GTC, GTG, and GTT, all of which are codons for valine. (Of course, the period matches any other character, but the $codon is assumed to have only A,C,G, or T characters.) The i after the regular expression means match uppercase or lowercase, for instance /T/i matches T or t. The new feature in these regular expressions is the use of the vertical bar or pipe (|) to separate two choices. Thus for serine, /TC.|AG[TC]/ matches /TC./ or /AG[TC]/. In this program, you need only two choices per regular expression, but you can use as many vertical bars as you like. You can also group parts of a regular expression in parentheses, and use vertical bars in them. For example, /give me a (break|meal)/ matches "give me a break" or "give me a meal." 8.3.4 Using Hashes for the Genetic CodeIf you think about using a hash for this translation, you'll see it's a natural way to proceed. For each codon key the amino acid value is returned. Here's the code: #
# codon2aa
#
# A subroutine to translate a DNA 3-character codon to an amino acid
# Version 3, using hash lookup
sub codon2aa {
my($codon) = @_;
$codon = uc $codon;
my(%genetic_code) = (
'TCA' => 'S', # Serine
'TCC' => 'S', # Serine
'TCG' => 'S', # Serine
'TCT' => 'S', # Serine
'TTC' => 'F', # Phenylalanine
'TTT' => 'F', # Phenylalanine
'TTA' => 'L', # Leucine
'TTG' => 'L', # Leucine
'TAC' => 'Y', # Tyrosine
'TAT' => 'Y', # Tyrosine
'TAA' => '_', # Stop
'TAG' => '_', # Stop
'TGC' => 'C', # Cysteine
'TGT' => 'C', # Cysteine
'TGA' => '_', # Stop
'TGG' => 'W', # Tryptophan
'CTA' => 'L', # Leucine
'CTC' => 'L', # Leucine
'CTG' => 'L', # Leucine
'CTT' => 'L', # Leucine
'CCA' => 'P', # Proline
'CCC' => 'P', # Proline
'CCG' => 'P', # Proline
'CCT' => 'P', # Proline
'CAC' => 'H', # Histidine
'CAT' => 'H', # Histidine
'CAA' => 'Q', # Glutamine
'CAG' => 'Q', # Glutamine
'CGA' => 'R', # Arginine
'CGC' => 'R', # Arginine
'CGG' => 'R', # Arginine
'CGT' => 'R', # Arginine
'ATA' => 'I', # Isoleucine
'ATC' => 'I', # Isoleucine
'ATT' => 'I', # Isoleucine
'ATG' => 'M', # Methionine
'ACA' => 'T', # Threonine
'ACC' => 'T', # Threonine
'ACG' => 'T', # Threonine
'ACT' => 'T', # Threonine
'AAC' => 'N', # Asparagine
'AAT' => 'N', # Asparagine
'AAA' => 'K', # Lysine
'AAG' => 'K', # Lysine
'AGC' => 'S', # Serine
'AGT' => 'S', # Serine
'AGA' => 'R', # Arginine
'AGG' => 'R', # Arginine
'GTA' => 'V', # Valine
'GTC' => 'V', # Valine
'GTG' => 'V', # Valine
'GTT' => 'V', # Valine
'GCA' => 'A', # Alanine
'GCC' => 'A', # Alanine
'GCG' => 'A', # Alanine
'GCT' => 'A', # Alanine
'GAC' => 'D', # Aspartic Acid
'GAT' => 'D', # Aspartic Acid
'GAA' => 'E', # Glutamic Acid
'GAG' => 'E', # Glutamic Acid
'GGA' => 'G', # Glycine
'GGC' => 'G', # Glycine
'GGG' => 'G', # Glycine
'GGT' => 'G', # Glycine
);
if(exists $genetic_code{$codon}) {
return $genetic_code{$codon};
}else{
print STDERR "Bad codon \"$codon\"!!\n";
exit;
}
}
This subroutine is simple: it initializes a hash and then performs a single lookup of its single argument in the hash. The hash has 64 keys, one for each codon. Notice there's a function exists that returns true if the key $codon exists in the hash. It's equivalent to the else statement in the two previous versions of the codon2aa subroutine.[3]
Also notice that to make this subroutine work on lowercase DNA as well as uppercase, you translate the incoming argument into uppercase to match the data in the %genetic_code hash. You can't give a regular expression to a hash as a key; it must be a simple scalar value, such as a string or a number, so the case translation must be done first. (Alternatively, you can make the hash twice as big.) Similarly, character classes don't work in the keys for hashes, so you have to specify each one of the 64 codons individually. You may wonder why bother wrapping this last bit of code in a subroutine at all. Why not just declare and initialize the hash and do the lookups directly in the hash instead of going through the subroutine? Well, the subroutine does do a little bit of error checking for nonexistent keys, so having a subroutine saves doing that error checking yourself each time you use the hash. Additionally, wrapping the code in a subroutine gives a little insurance for the future. If all the code you write does codon translation by means of our subroutine, it would be simplicity itself to switch over to a new way of doing the translation. Perhaps a new kind of datatype will be added to Perl in the future, or perhaps you want to do lookups from a database or a DBM file. Then all you have to do is change the internals of this one subroutine. As long as the interface to the subroutine remains the same—that is to say, as long as it still takes one codon as an argument and returns a one-character amino acid—you don't need to worry about how it accomplishes the translation from the standpoint of the rest of the programs. Our subroutine has become a black box. This is one significant benefit of modularization and organization of programs with subroutines. There's another good, and biological, reason why you should use a subroutine for the genetic code. There is actually more than one genetic code, because there are differences as to how DNA encodes amino acids among mammals, plants, insects, and yeast—especially in the mitochondria. So if you have modularized the genetic code, you can easily modify your program to work with a range of organisms. One of the benefits of hashes is that they are fast. Unfortunately, our subroutine declares the whole hash each time the subroutine is called, even for one lookup. This isn't so efficient; in fact, it's kind of slow. There are other, much faster ways that involve declaring the genetic code hash only once as a global variable, but they would take us a little far afield at this point. Our current version has the advantage of being easy to read. So, let's be officially happy with the hash version of codon2aa and put it into our module in the file BeginPerlBioinfo.pm (see Chapter 6). Now that we've got a satisfactory way to translate codons to amino acids, we'll start to use it in the next section and in the examples.
|
Index terms contained in this section() (parentheses)grouping in regular expressions | (vertical bar) regular expressions, use in amino acids translating DNA into representing with codons biology molecular central dogma central dogma of molecular biology character classes genetic code, redundancy of hash keys and codons amino acids, representing with translating to amino acids testing for redundant codons defined function DNA translating into amino acids and proteins error messages directing to STDERR exists function genetic code hashes, using for redundancy in 2nd translating codons to amino acids hashes genetic code, using for input STDIN (standard input) molecular biology central dogma of nucleotides codons output directing with STDOUT patterns (and regular expressions) choices, separating with vertical bar (|) genetic code, redundancy in hash keys and printing to STDOUT proteins DNA, translating into RNA, translation into redundancy in genetic code 2nd RNA (ribonucleic acid) DNA, transcribing to stop signals subroutines redundant codons, testing for translating DNA codons to amino acids testing for redundant codons using hash lookup transcription translation DNA codons to amino acids redundant codons, testing for using hash lookup |