 4.4 Transcription: DNA to RNAA large part of what you, the Perl bioinformatics programmer, will spend your time doing amounts to variations on the same theme as Examples 4-1 and 4-2. You'll get some data, be it DNA, proteins, GenBank entries, or what have you; you'll manipulate the data; and you'll print out some results. Example 4-3 is another program that manipulates DNA; it transcribes DNA to RNA. In the cell, this transcription of DNA to RNA is the outcome of the workings of a delicate, complex, and error-correcting molecular machinery.[3] Here it's a simple substitution. When DNA is transcribed to RNA, all the T's are changed to U's, and that's all that our program needs to know.[4]
Example 4-3. Transcribing DNA into RNA#!/usr/bin/perl -w # Transcribing DNA into RNA # The DNA $DNA = 'ACGGGAGGACGGGAAAATTACTACGGCATTAGC'; # Print the DNA onto the screen print "Here is the starting DNA:\n\n"; print "$DNA\n\n"; # Transcribe the DNA to RNA by substituting all T's with U's. $RNA = $DNA; $RNA =~ s/T/U/g; # Print the RNA onto the screen print "Here is the result of transcribing the DNA to RNA:\n\n"; print "$RNA\n"; # Exit the program. exit; Here's the output of Example 4-3: Here is the starting DNA: ACGGGAGGACGGGAAAATTACTACGGCATTAGC Here is the result of transcribing the DNA to RNA: ACGGGAGGACGGGAAAAUUACUACGGCAUUAGC This short program introduces an important part of Perl: the ability to easily manipulate text data such as a string of DNA. The manipulations can be of many different sorts: translation, reversal, substitution, deletions, reordering, and so on. This facility of Perl is one of the main reasons for its success in bioinformatics and among programmers in general. First, the program makes a copy of the DNA, placing it in a variable called $RNA: $RNA = $DNA; Note that after this statement is executed, there's a variable called $RNA that actually contains DNA.[5] Remember this is perfectly legal—you can call variables anything you like—but it is potentially confusing to have inaccurate variable names. Now in this case, the copy is preceded with informative comments and followed immediately with a statement that indeed causes the variable $RNA to contain RNA, so it's all right. Here's a way to prevent $RNA from containing anything except RNA:
($RNA = $DNA) =~ s/T/U/g; In Example 4-3, the transcription happens in this statement: $RNA =~ s/T/U/g; There are two new items in this statement: the binding operator (=~) and the substitute command s/T/U/g. The binding operator =~ is used, obviously enough, on variables containing strings; here the variable $RNA contains DNA sequence data. The binding operator means "apply the operation on the right to the string in the variable on the left." The substitution operator , shown in Figure 4-1, requires a little more explanation. The different parts of the command are separated (or delimited) by the forward slash. First, the s indicates this is a substitution. After the first / comes a T, which represents the element in the string that will be substituted. After the second / comes a U, which represents the element that's going to replace the T. Finally, after the third / comes g. This g stands for "global" and is one of several possible modifiers that can appear in this part of the statement. Global means "make this substitution throughout the entire string," that is to say, everywhere possible in the string. Figure 4-1. The substitution operator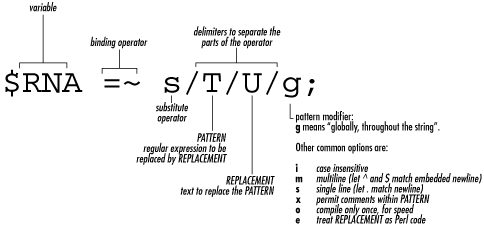 Thus, the meaning of the statement is: "substitute all T's for U's in the string data stored in the variable $RNA." The substitution operator is an example of the use of regular expressions. Regular expressions are the key to text manipulation, one of the most powerful features of Perl as you'll see in later chapters.
|
Index terms contained in this section/ (forward slash)s/// (substitution) operator = (equal sign) =~ (pattern binding) operator assignment in DNA to RNA transcription binding operators (=~) DNA transcribing to RNA global substitution operators binding s/// (substitution) patterns (and regular expressions) substitution operator Perl text data, ease of manipulating regular expressions reverse complements DNA transcription to RNA RNA (ribonucleic acid) transcribing DNA to substitution (s///) operator text Perl, ease of manipulating thymine (T) replacing with uracil (U) in DNA to RNA transcription transcription DNA to RNA uracil (U) replacing thymine (T) in DNA to RNA transcription variables containing strings, using binding operator (=~) with |